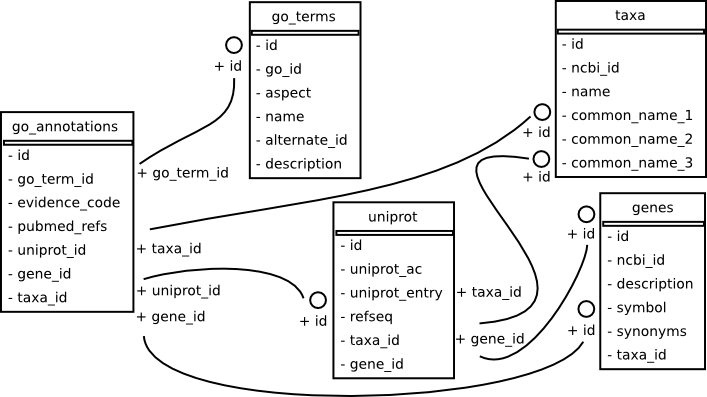
Database¶
The schema for the database.
The main uses of the database are documented here:
Setting up your database¶
There are 4 step that you need to carry out in order to have a working database.
- Install PostgreSQL and create an empty database
- Modify the htsint config file
- Fetch all necessary data files
- Populate the datbase
(1) PostgreSQL setup¶
Install PostgreSQL on you system. For example on Ubuntu
~$ sudo apt-get install postgresql
Then you need to create a database. Here we do this with a database named htsintegrate, a user named dbuser and a password somepassword. You should change these parameters to fit your system.
~$ sudo su - postgres ~$ psql -U postgres CREATE USER dbuser WITH ENCRYPTED PASSWORD 'somepassword'; CREATE DATABASE htsintegrate WITH OWNER dbuser; \q
Visit the PostgreSQL documentation to learn more.
(2) Modify the config file¶
Set the database name, user, password and port
>>> from htsint import Configure
>>> config = Configure()
>>> config.log['dbuser'] = 'dbuser'
>>> config.log['dbname'] = 'htsintegrate'
>>> config.log['dbpass'] = 'somepassword'
>>> config.save()
The other variables than can be modified are:
>>> print(config.log.keys())
['dbport', 'taxa', 'dbhost', 'dbpass', 'dbuser', 'blast', 'data', 'dbname']
Check that all taxa that you would like annotation information for are present. You can print the default taxa with
>>> print(config.log['taxa'])
The default taxa are:
3702, Arabidopsis thaliana 4932, Saccharomyces cerevisiae 5476, Candida albicans 7227, Drosophila melanogaster 7955, Danio rerio 8355, Xenopus laevis 8364, Xenopus (Silurana) tropicalis 9031, Gallus gallus 9606, Homo sapiens 10090, Mus musculus 10566, Human papillomavirus 10116, Rattus norvegicus 28377, Anolis carolinensis
If you would like to add, for example, Solanum lycopersicum then use typical list syntax. You need to save any changes you make.
>>> config.log['taxa'].append('4081')
>>> config.save()
There is a good chance you will want to specify the directory where all the downloaded data is stored. This can be done with any valid full path.
The default is
>>> print(config.log['data'])
'/usr/local/share/htsint'
Alternatively, it is possible to edit this file directly. To locate the directory where it is stored you may type the following.
>>> import os
>>> os.path.join(os.path.expanduser('~'),".htsint")
'/home/adam/.htsint'
The dbport (default ‘5432’) and dbhost (default ‘localhost’) may also be configured.
Note
htsint will only populate annotation information for taxa in the taxa variable so make sure all species are present before database population.
(3) Fetch the necessary data files¶
The rest of the process is automated assuming you have created your PostgreSQL database and modified you config file.
>>> from htsint.database import DatabaseFetch
>>> fetch = DatabaseFetch()
>>> fetch.run()
This class only currently works under Linux/OSX operating systems. For other systems the following files could be downloaded by hand and placed in the ‘data’ directory.
The fetching can take several hours depending on the speed of your connection. The compressed files total less than 15GB, but be aware that the uncompressed versions will take up over 100GB of space. If space is an issue all files may be erased except uniprot_sprot.fasta.* and go.obo as the former is used for BLAST and the latter is not stored directly in the database and is used as part of most analysis pipelines.
A logfile is produced and stored in your data directory.
(4) Populate the database¶
Finally, the database can be populated with the following class.
>>> from htsint.database import DatabaseCreate
>>> db = DatabaseCreate()
>>> db.run()
A typical database will take a little over an hour to populate. A logfile is produced and stored in your data directory.
A summary can be produced at any time using which will produce a similar output.
>>> from htsint.database import print_db_summary
>>> print_db_summary()
DATABASE - htsintegrate - SUMMARY There are 1262260 entries in the taxa table There are 681732 entries in the genes table There are 777608 entries in the uniprot table There are 42627 entries in the go_terms table There are 7463568 entries in the go_annotations table
Additional Notes¶
What exactly is stored in the database?¶
- All taxa from NCBI taxonomy
- Gene, UniProt and GO annotation information for only the specified taxa
- All information about GO terms
Database updating¶
Because of the challenges that can arise through naming conflicts when updating NCBI and UniProt data it is recommended that you run the fetch and create steps again, which will create a clean updated version. The fetch step will check if a current file is the newest and only download a new one if necessary.
Database portability¶
You may want to copy your database to another computer instead of waiting for the db to populate. This can be done as follows.
To create a file that may be transferred to another computer
~$ pg_dump -h localhost -U dbuser dbname > htsint.sql
To add the database to another server
~$ sudo su - postgres ~$ psql -U postgres CREATE USER dbuser WITH ENCRYPTED PASSWORD 'somepassword'; CREATE DATABASE newdbname WITH OWNER dbuser; \q ~$ psql newdbname < htsint.sql
See the database cookbook for more information on getting started with the database.