Voraphani 2014¶
Basic info¶
- GEO: GSE43696
- reference: [Voraphani14]
- platform: Agilent Human GE 4x44K V2
- tissue: bronch biopsy
- tissue environment: ex-vivo
Note
The asthma dx was specified as severe and moderate so all non-control dxs were set to asthma.
Samples¶
| asthma | AR | controls | total |
|---|---|---|---|
| 66 | 0 | 25 | 91 |
The following is a summary of the covariate information extracted from downloaded GSE files.
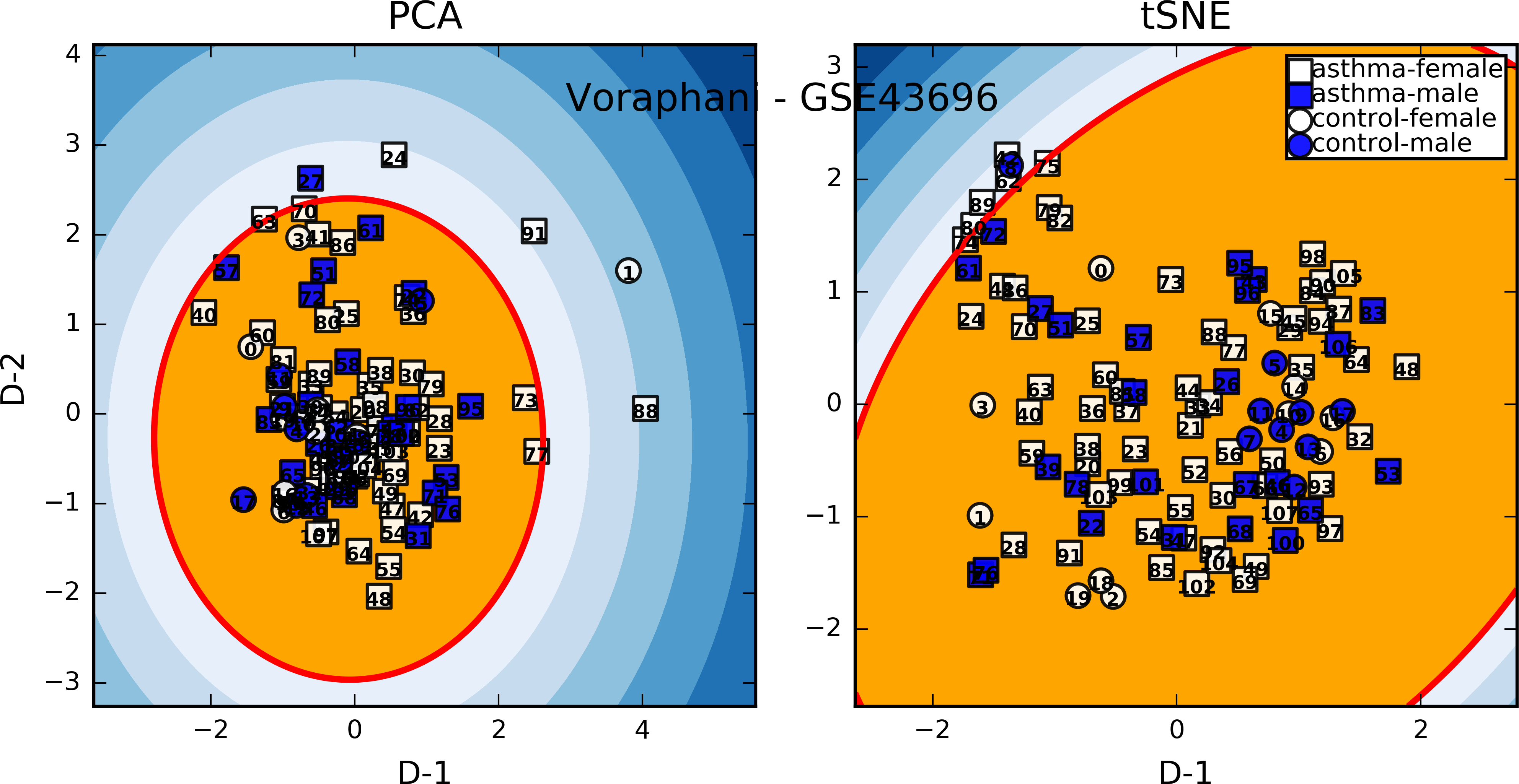
Gene expression analysis¶
Tue Aug 09 08:47:20 AM 2016
total files: 1
file: 1/1
file name: GSE43696_series_matrix.txt.gz
status: Public on Jul 01 2014
data processing: Agilent Feature Extraction v10.7.3.1 default parameters
type: RNA
probes 41000
samples 108
platform_id: GPL6480
covariates extracted from eset...
disease_state
cell_type
gender
age
model: ~0+dx+age+gender
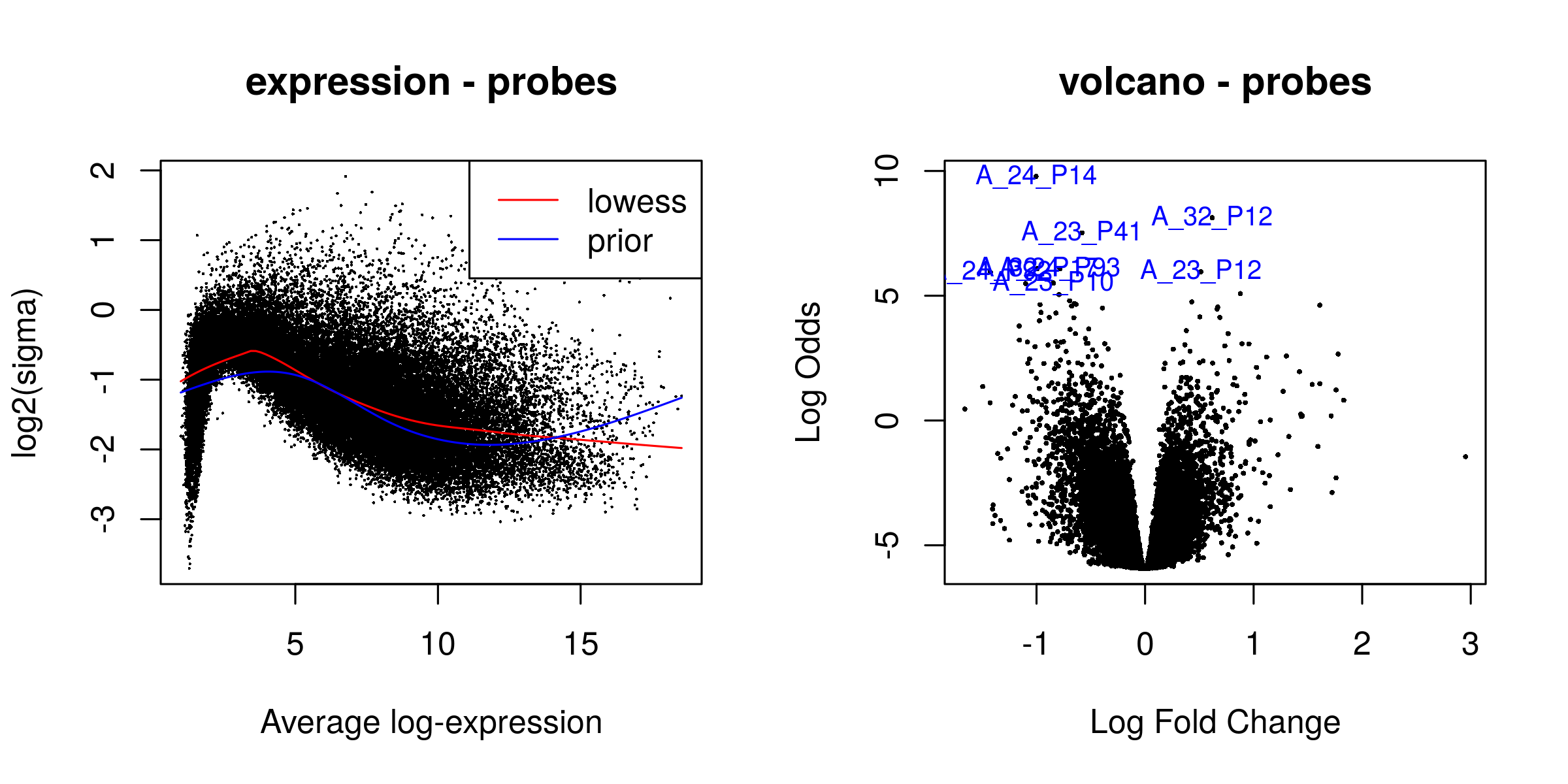
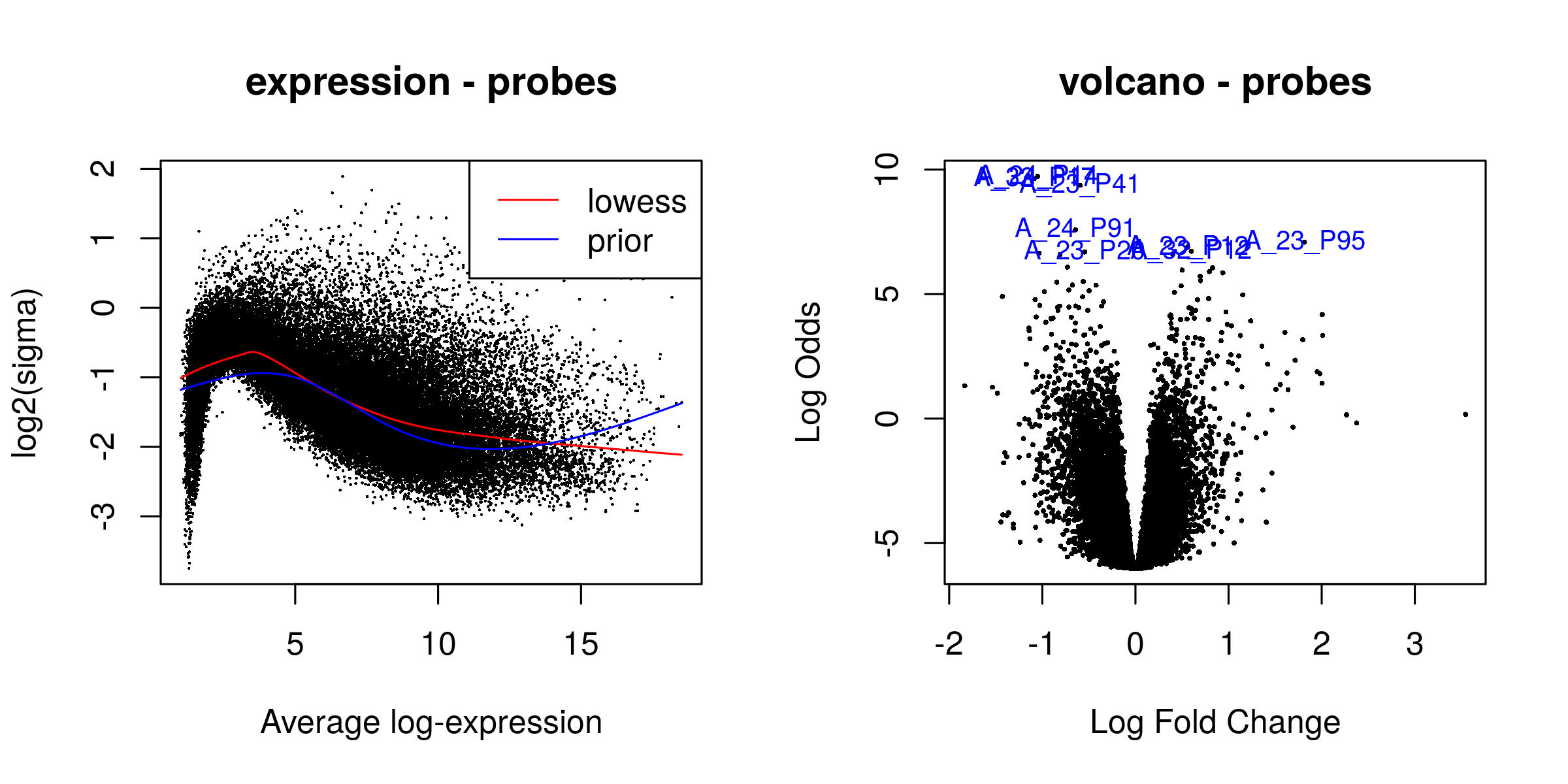
gene filter 30936/41000
There were 268 significant probes
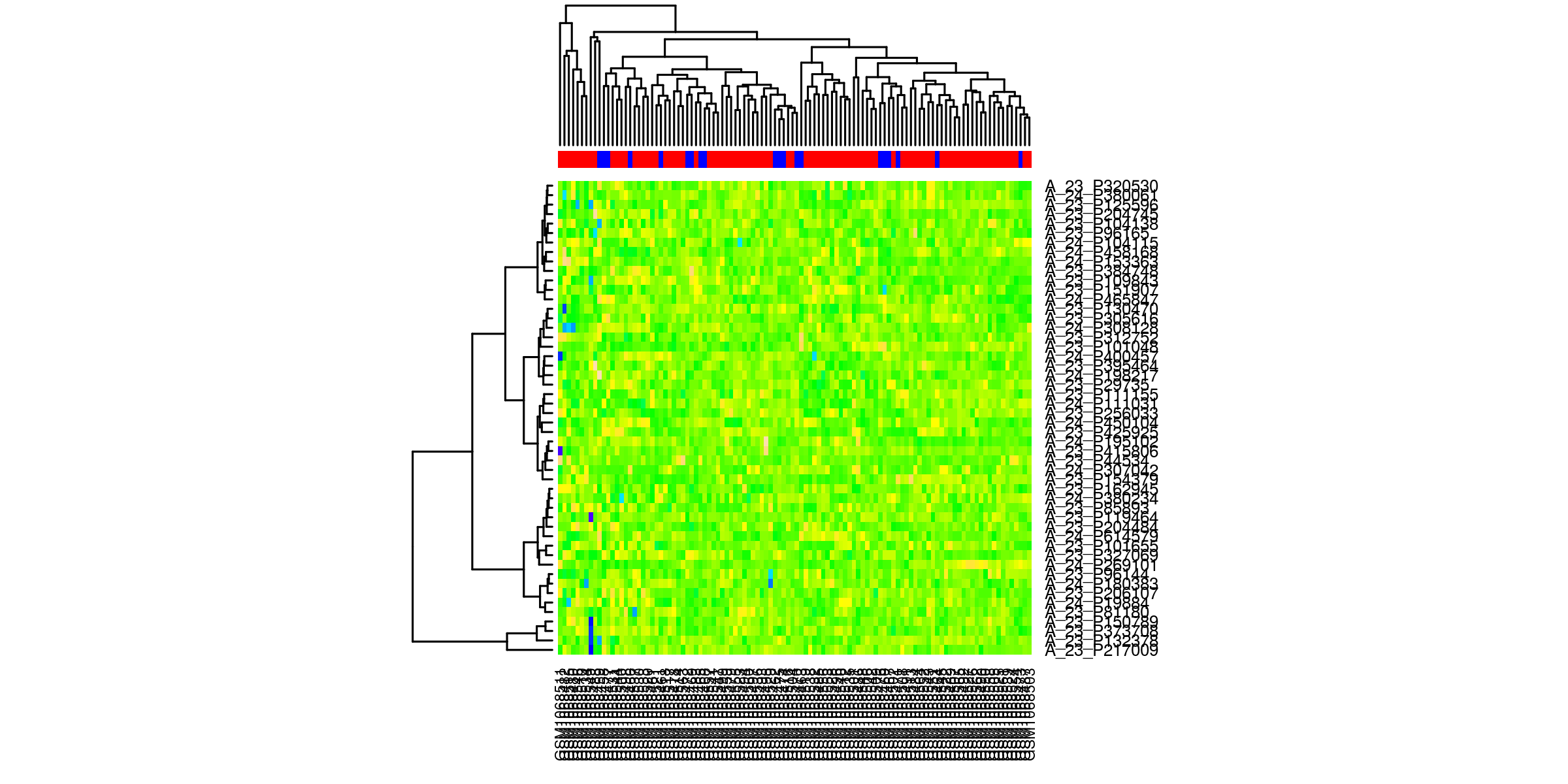
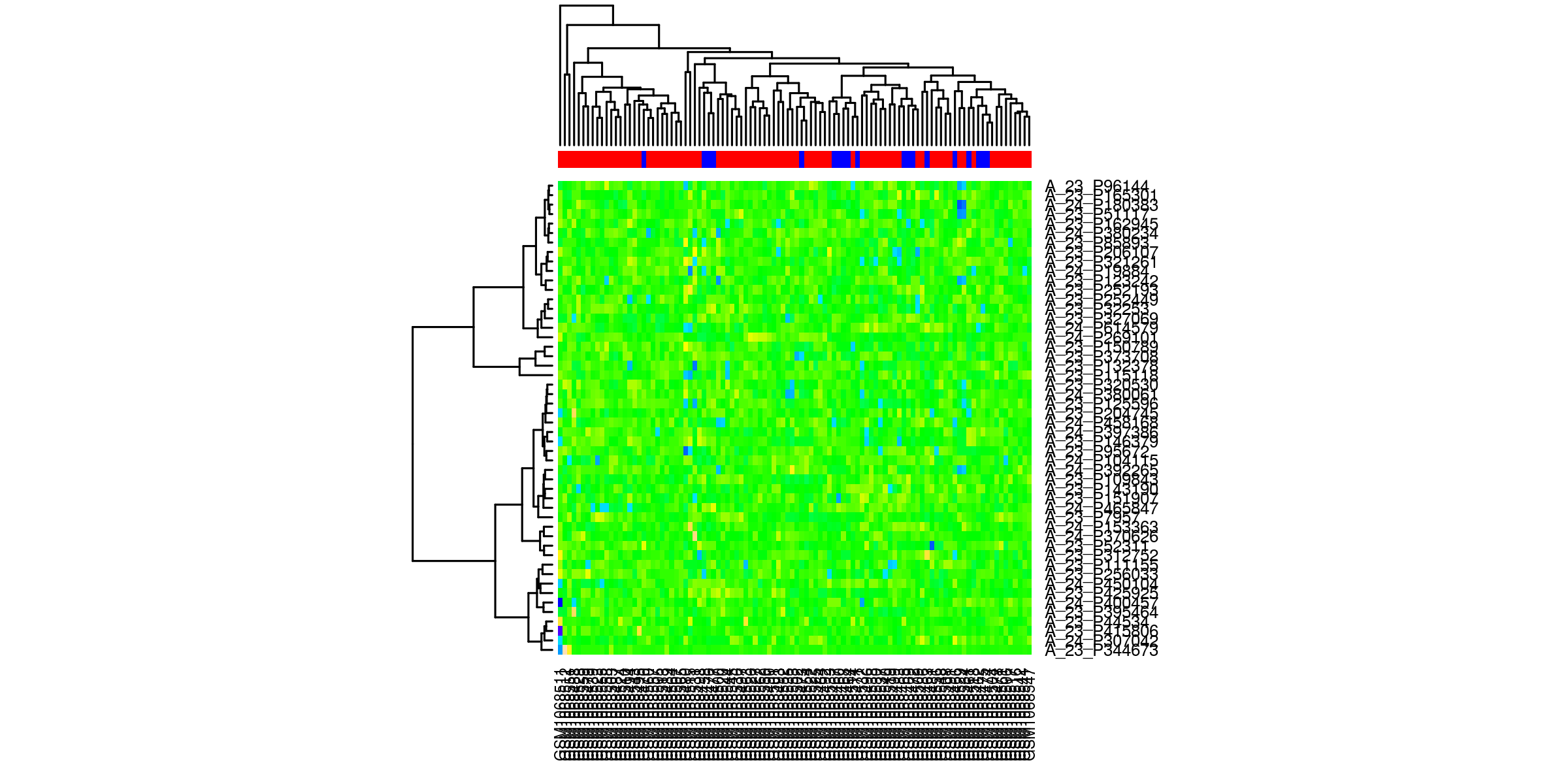
Heatmaps¶
Outliers removed¶
Heatmaps of the top 50 genes. The genes are clustered based on normalized values of intensity. The quality control check is that the samples cluster according to phenotype.