Christenson 2015¶
Basic info¶
- GEO: GSE67472
- reference: unpublished
- platform: Affymetrix Human Genome U133 Plus 2.0 Array
- tissue: bronch biopsy
- tissue environment: ex-vivo
Samples¶
| asthma | AR | controls | total |
|---|---|---|---|
| 62 | 0 | 43 | 105 |
Gene expression analysis¶
Tue Aug 09 08:46:38 AM 2016
total files: 1
file: 1/1
file name: GSE67472_series_matrix.txt.gz
status: Public on Apr 01 2015
data processing: Data underwent RMA normalization and Combat batch correction. Probeset IDs were defined using the Entrez Gene custom CDF v14 available for the Affymetrix U133 Plus 2.0 array available at brainarray.mbni.med.umich.edu.
type: RNA
probes 19008
samples 105
platform_id: GPL16311
covariates extracted from eset...
tissue
age
gender
disease_state
th2_group
model: ~0+dx+age+gender+batch+race
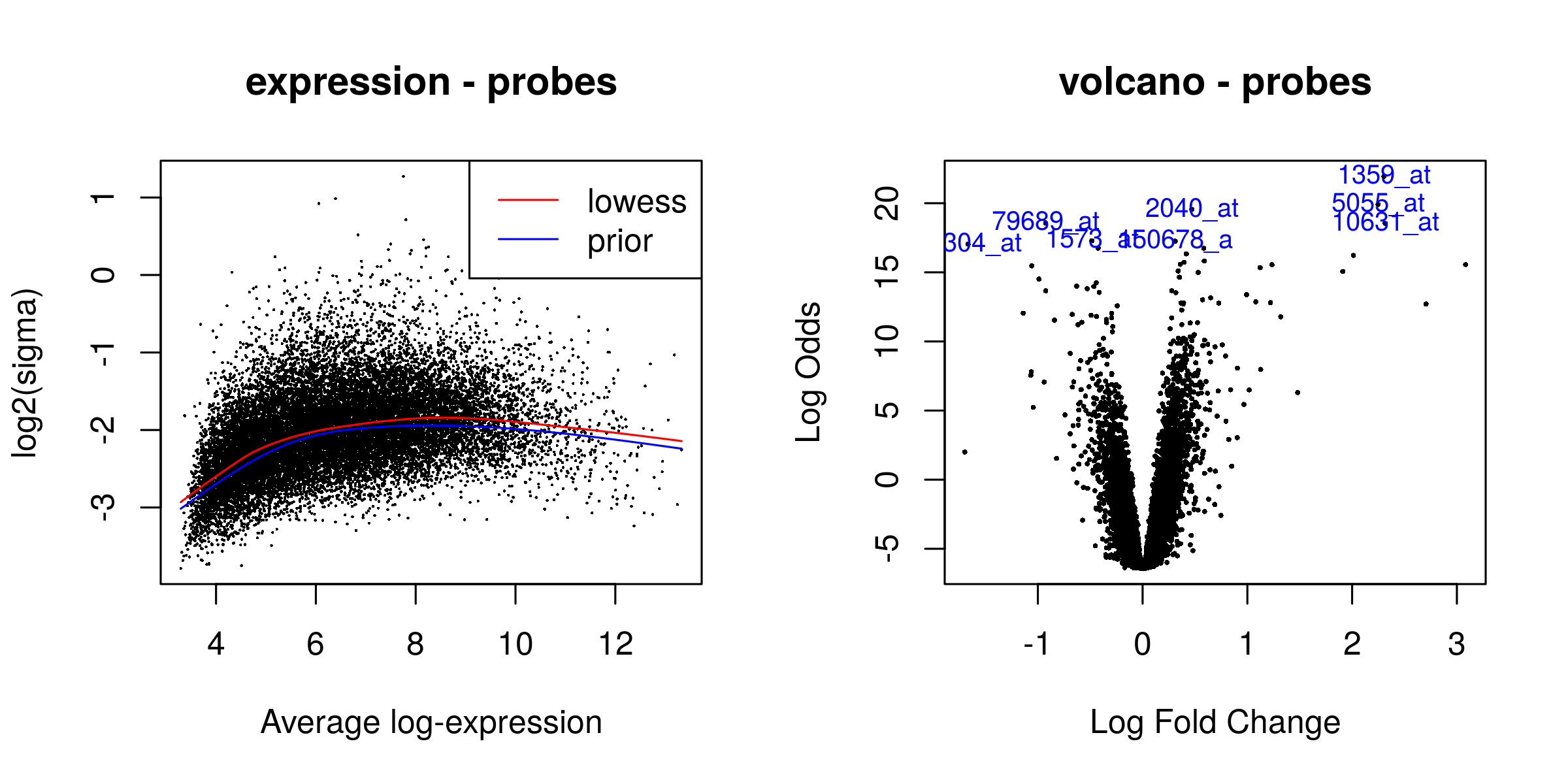
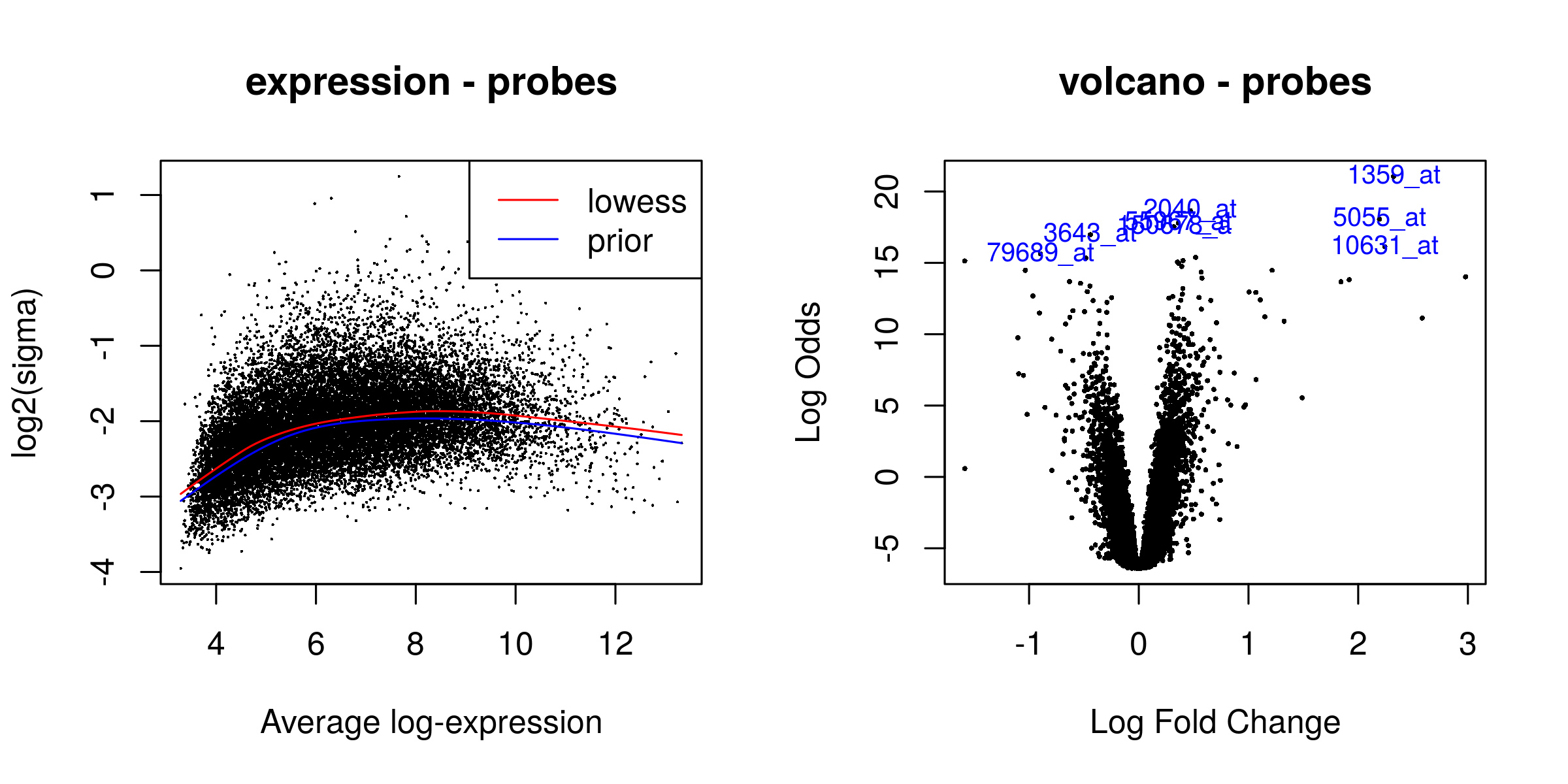
gene filter 19008/19008
There were 3566 significant probes
Notes¶
They used a custom CDF file to define probeset IDs. For more information you can read about it here. It says that they used version 14 (March, 2011) so I am a little confused, because the purpose of redefining the probesets in the first place is because the mapping is a bit behind UniGene, so it is odd they are using such an old version if they are concerned with lag.
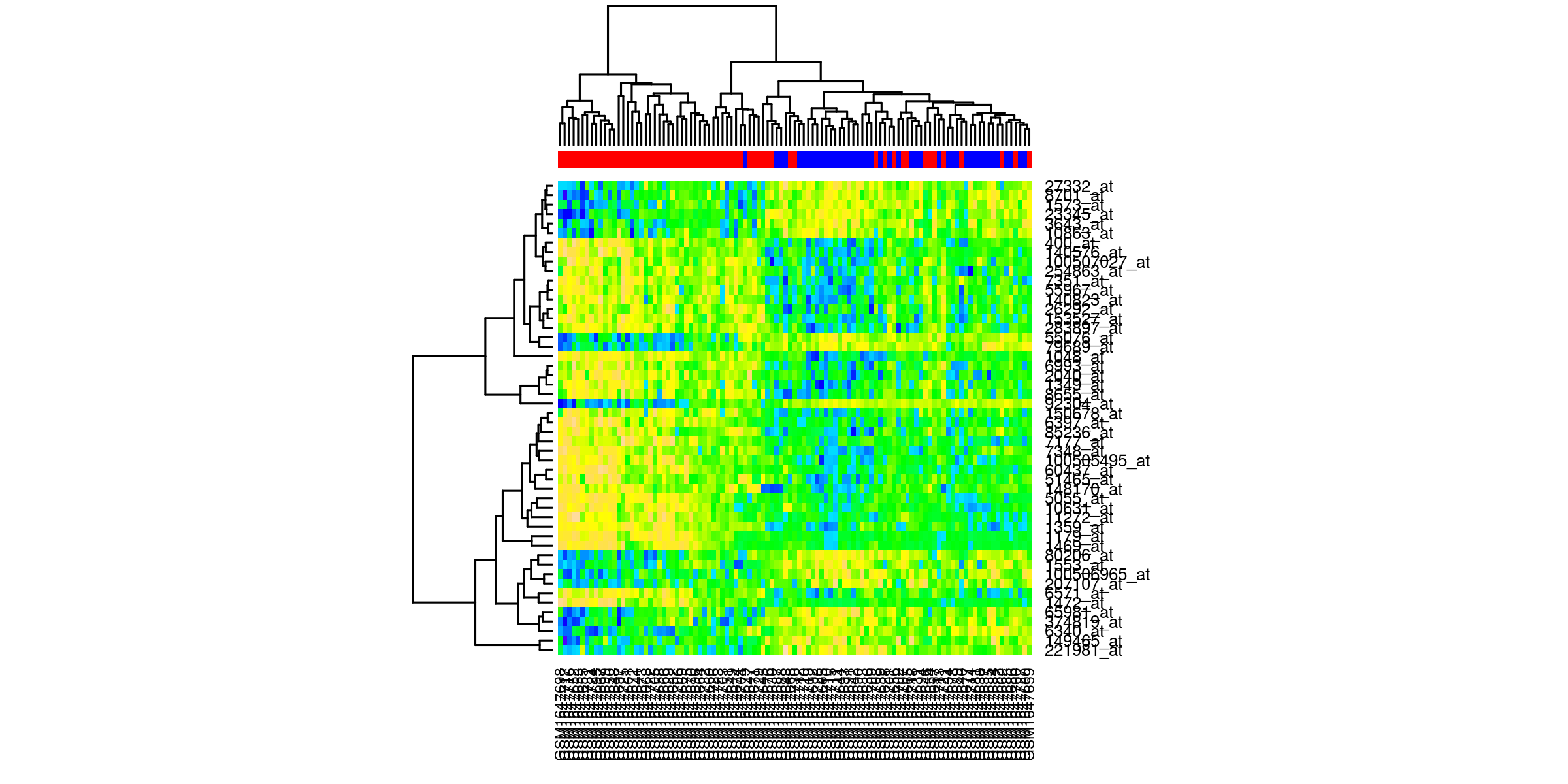
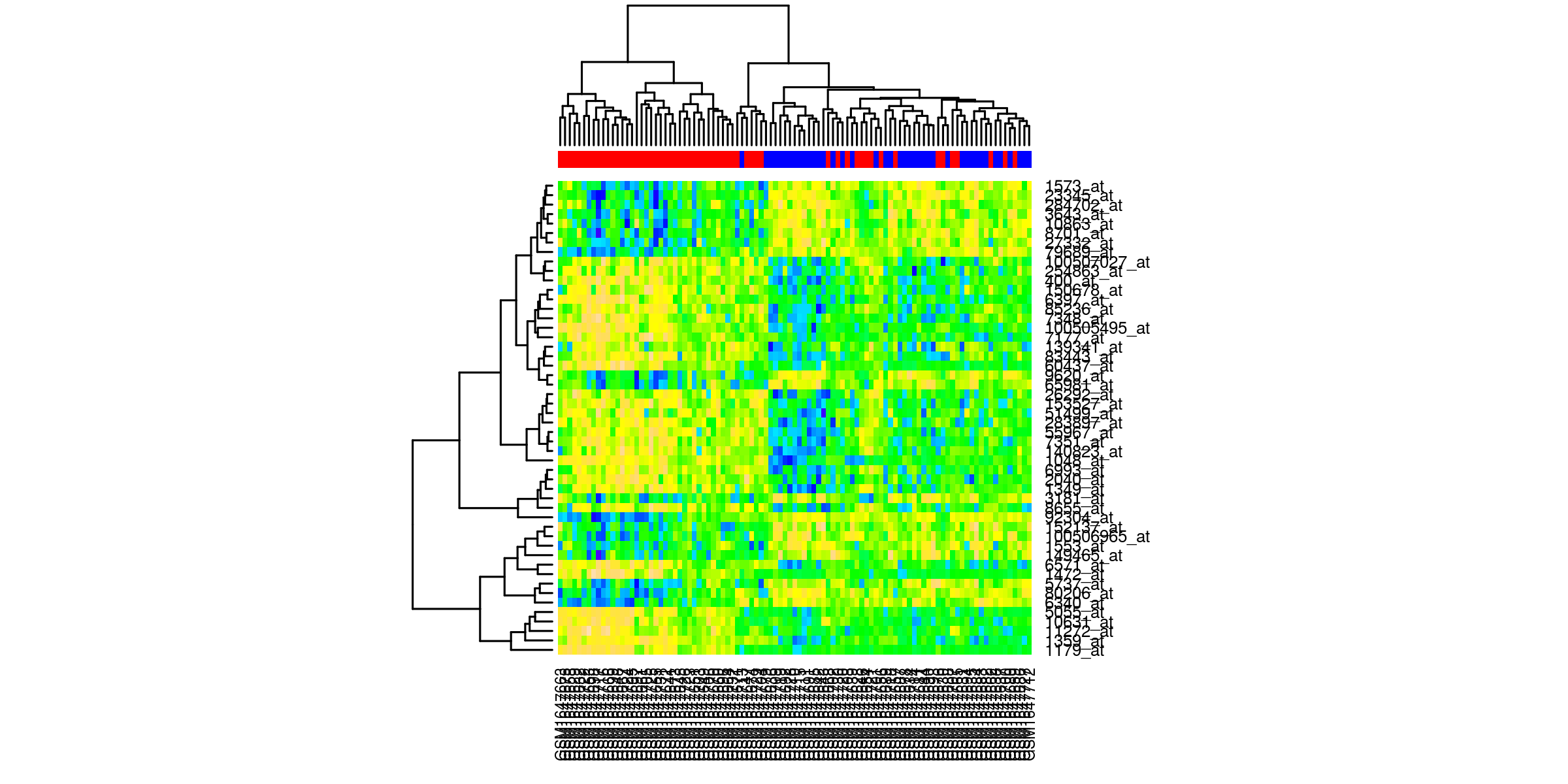
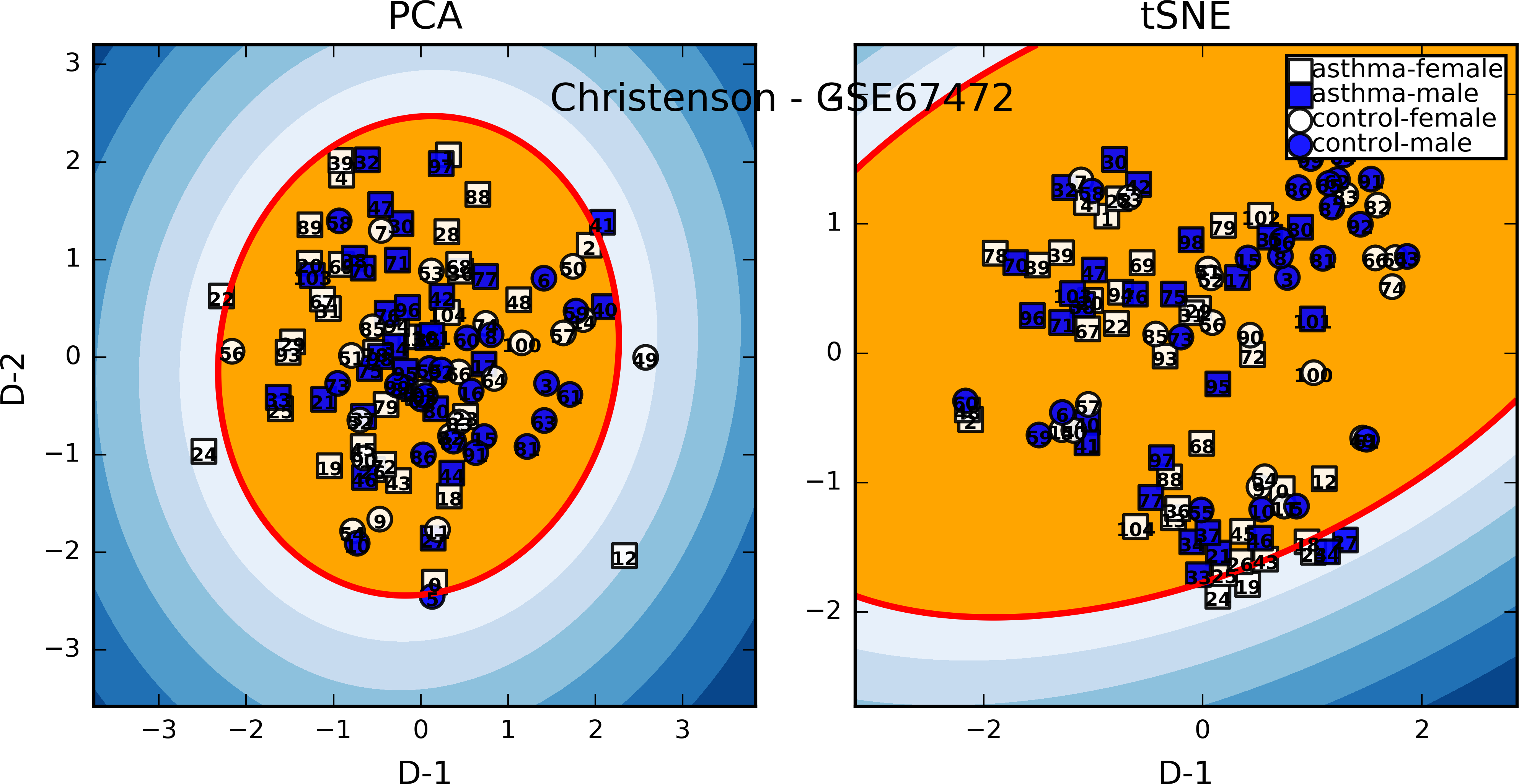
Heatmaps¶
Outliers removed¶
Heatmaps of the top 50 genes. The genes are clustered based on normalized values of intensity. The quality control check is that the samples cluster according to phenotype.