Database¶
About the database¶
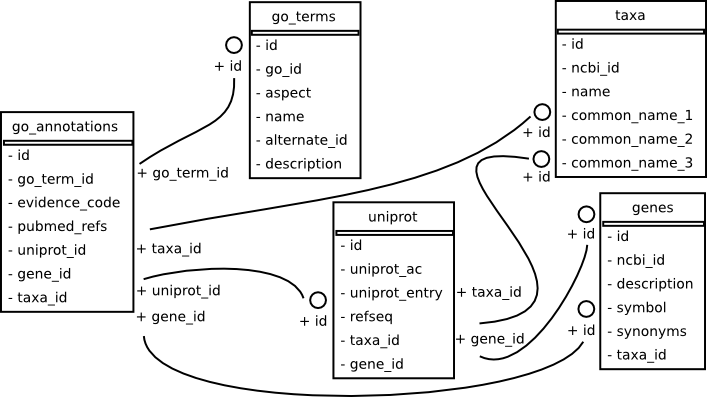
The databased we used for this analysis used PostgreSQL and several python modules, including the object relational mapper provided by SQLAlchemy. The main purpose of the database is to be able to handle at a detailed level Gene Ontology [Ashburner00] information. Here is the relational schema produced with a tool called sqlalchemy_schemadisplay. The database population scripts along with a number of classes to interface with the data are made possible through a project called hts-integrate.
Database contents¶
The following files were downloaded on September 15, 2014.
The uniprot and gene centric data from these files was parsed and used to populate the database. The number of rows in each of the tables are shown below.
There are 1262260 entries in the taxa table There are 681732 entries in the genes table There are 777608 entries in the uniprot table There are 42627 entries in the go_terms table There are 7463568 entries in the go_annotations table